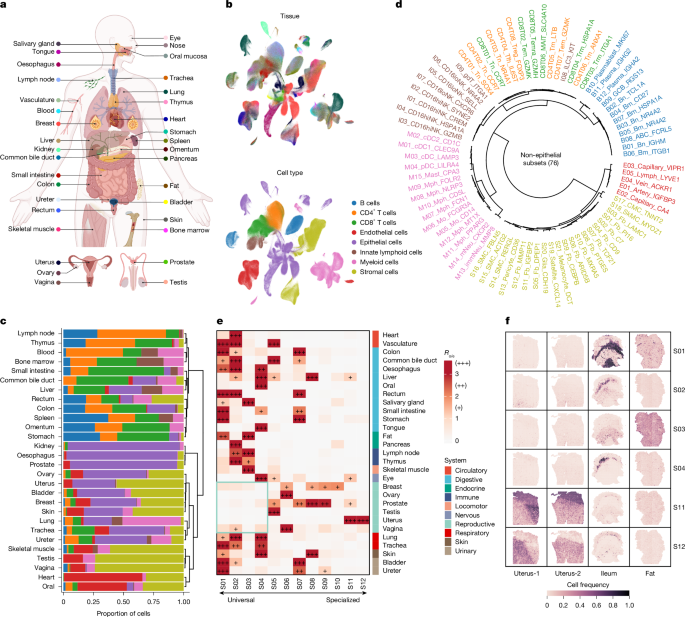
"We collected scRNA-seq datasets from healthy samples to minimize batch effects and developed a comprehensive pan-tissue cell atlas representing 35 human tissues."
"Quality control was implemented using the Scanpy toolkit, filtering datasets for specific criteria to ensure the reliability and consistency of single-cell RNA sequencing data."
"Our stringent preprocessing methodology involved removing low-quality cells and maintaining only high-quality cells, resulting in over 700 samples that successfully met quality assurance."
"By regulating the collection criteria and quality standards, we achieved a standardized transcriptome across different datasets, ensuring high accuracy for the pan-tissue cell atlas."
This article outlines the process of assembling a pan-tissue cell atlas through the collection and preprocessing of single-cell RNA sequencing (scRNA-seq) datasets from healthy adult samples. A total of 33 datasets were included, chosen based on specific criteria that aimed to minimize batch effects. The quality control involved using Scanpy for data standardization, with strict parameters guiding the selection of high-quality cells, resulting in a consolidated dataset representing 35 human tissues. This methodology ensures the reliability and comprehensiveness of the resultant cell atlas.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]