"Therapeutic genome-editing efforts, including more than 70 clinical trials so far, have predominantly used programmable nucleases, base editors or prime editors to disrupt or correct disease-associated genes in an allele-specific manner. These approaches have proven to be effective in patients or in animal models for the treatment of disorders such as sickle-cell disease6,7, T cell leukaemia8, hypercholesterolaemia9,10, alpha-1-antitrypsin deficiency10, chronic granulomatous disease11, progeria12, spinal muscular atrophy13, prion disease14, alternating hemiplaegia of childhood15 and many other genetic diseases. Although allele-specific therapeutic genome-editing strategies offer treatments for many serious diseases with few treatment options, the breadth of the global genetic disease crisis,"
"Allele-specific applications of nucleases, base editors and prime editors require the development of a distinct genome-editing treatment for each of the more than 200,000 known pathogenic mutations, although prime editing also enables corrections of clustered hotspots of mutations. Until substantial streamlining of regulatory, development and manufacturing costs occurs, the development of the thousands of distinct drugs needed to treat any large fraction of patients with genetic disease remains impractical, despite the fact that base editing and prime editing can collectively correct the vast majority of known pathogenic mutations."
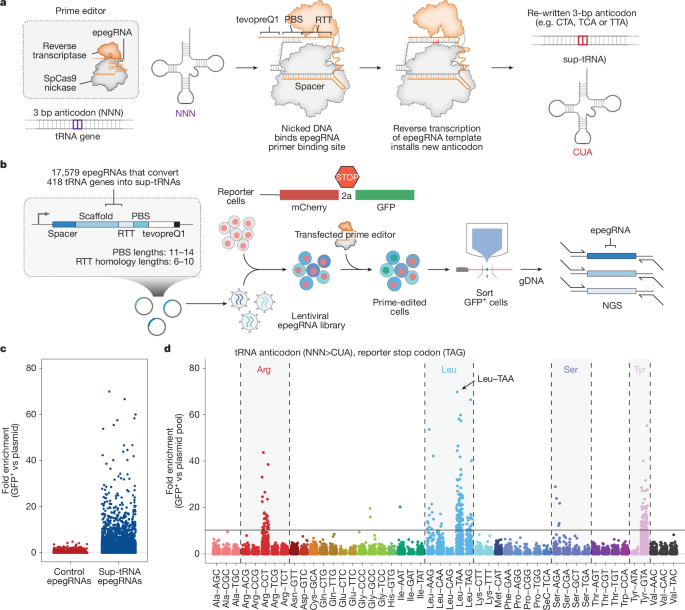
More than 70 clinical trials have used programmable nucleases, base editors or prime editors to disrupt or correct disease-associated genes in an allele-specific manner and have shown effectiveness in patients or animal models across many genetic disorders. The global burden of over 8,000 genetic diseases affecting hundreds of millions of patients requires approaches that serve large numbers of patients more rapidly. Allele-specific strategies necessitate distinct treatments for over 200,000 pathogenic mutations, making broad deployment impractical without regulatory and manufacturing streamlining. Prime editing, using a programmable nickase and reverse transcriptase to replace targeted DNA segments, offers versatile opportunities to benefit larger patient groups. Nonsense mutations account for 24% of pathogenic alleles.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]