"The growing interest in bacteriophages is driven by the richness of unexplored genes that emerge in the phage-host conflict1,2. Targeted mapping and characterization of these genes is key to understanding a phage's infection cycle and its ability to counter host defences, but the diversity and genetic intractability of many phages and their hosts pose major challenges. Phage functional genomics approaches have used targeted inhibition of viral gene expression through, for example, small RNAs3,4 or CRISPR-Cas technology."
"However, the limited genetic tractability of many phage-host pairs due to defence systems that target foreign DNA1,2 or phage-mediated neutralization of Cas enzymes8,9 remains a barrier, even for intensely studied phages such as the model jumbo phage ΦKZ of the major human pathogen Pseudomonas aeruginosa. ΦKZ-like phages are of interest not only because of their potential for phage therapy of recalcitrant infections10, but also owing to their complex infection cycle, which features the sequential formation of membrane- and protein-bound compartments."
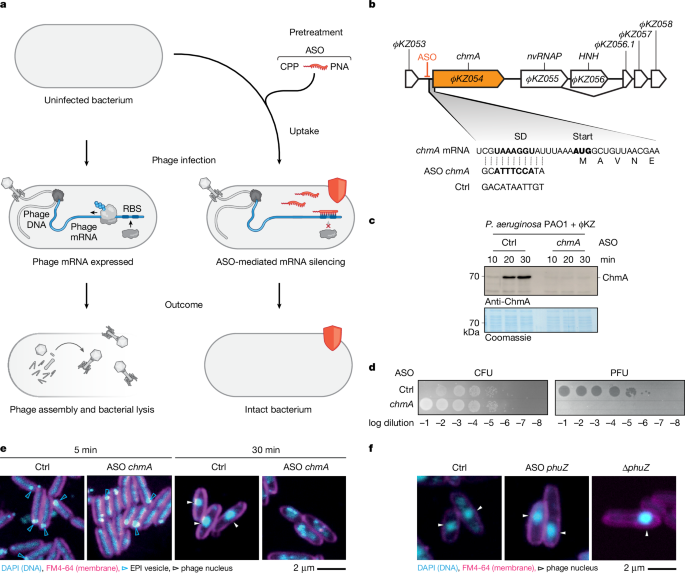
"ΦKZ injects its 280-kb dsDNA genome together with a virion RNA polymerase (vRNAP) in an early phage infection (EPI) vesicle for immediate phage gene transcription. Subsequently, a 'phage nucleus' forms, in which the non-virion RNA polymerase (nvRNAP) continues transcription14,15. The phage genome is then replicated and loaded into phage capsids (Extended Data Fig. 1a). Several conserved phage factors enable the formation and organization of the phage nucleus-for example, the nuclear shell protein chimallin (ChmA), PicA (also known as Imp1), which mediates cargo trafficking into the phage nucleus18,19, and the tubulin-like protein PhuZ, which centres the phage nucleus in the middle of the cell and helps to traffic newly assembled capsids."
Bacteriophage research is driven by numerous unexplored genes arising from phage-host conflicts. Mapping and characterizing these genes is essential to reveal infection-cycle mechanisms and counterdefence strategies, but phage and host diversity and genetic intractability present major obstacles. Functional genomics methods such as small RNAs and CRISPR-Cas have been applied, yet host defence systems and phage neutralization of Cas proteins limit their effectiveness. The jumbo phage ΦKZ exemplifies complexity with compartmentalized infection stages: EPI vesicles with vRNAP, a phage nucleus with nvRNAP, genome replication and capsid loading, and conserved factors like ChmA, PicA and PhuZ organizing the nucleus.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]