"These viruses are thought to shape the gut microbial community through predation, horizontal gene transfer and lysogenic conversion6,7. Recent advances in computational mining of gut metagenomes have revealed an expansive collection of viral metagenome-assembled genomes and efforts cataloguing this diversity have led to the discovery of several important viral families1,2,3,8,9,10. Moreover, lysogeny is common within the gut, with up to 90% of bacteria predicted to harbour prophages11,12."
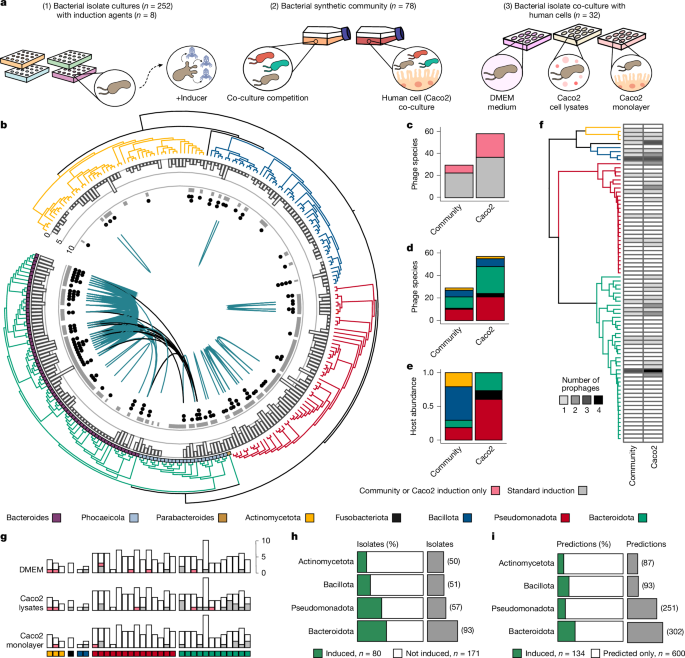
"Here we use a collection of 252 human gut bacterial isolates (50 Actinomycetota, 1 Fusobacteriota, 51 Bacillota, 57 Pseudomonadota and 93 Bacteroidota) to computationally identify and experimentally validate inducible prophages (Fig. 1a and Supplementary Table 1). We began by exposing our bacterial isolate cultures to eight different induction agents and conditions, which included a standard medium control, well-known inducing agents such as mitomycin C (0.3 and 3 µg ml −1)"
The human gut contains numerous microorganisms and viruses, including phages that shape microbial communities via predation, horizontal gene transfer and lysogenic conversion. Lysogeny is common in the gut, with up to 90% of bacteria predicted to harbour prophages, but the frequency and triggers of prophage re-entry into lytic replication remain unclear. Inactivation of resident prophages enables bacterial populations to escape lysis while retaining beneficial phage genes. Initiation of lytic replication depends on both host- and phage-specific cues. A set of 252 human gut bacterial isolates across five phyla was used to computationally identify and experimentally validate inducible prophages by testing eight induction agents and conditions.
Read at Nature
Unable to calculate read time
Collection
[
|
...
]